Abstract
Background and aims
The amount of clinical and biological data stored within clinical trials is growing exponentially. Data warehousing (DW) is useful for systematic global evaluation of information collected in trials: the highly translational FIL(Fondazione Italiana Linfomi)-MCL0208 trial has been used to test DW to improve data quality and to discover putative associations [Zaccaria, ASH 17].
In this study we developed an engineered prognostic model, focusing on easily accessible clinical variables. For this purpose, we exploited hierarchical clustering with the aim of seeking hidden patterns of interest in large datasets. Hence, these tools allowed to develop a novel prognostic model: the engineered MIPI index (e-MIPI).
Herein we present the first results, on baseline clinical characteristics:
clustering analysis and definition of a signature of predictive variables
construction of the e-MIPI to detect patients' risk of relapse
comparison with known prognostic indexes for MCL
validation of the signature on independent subset of patients.
Methods
Data were retrieved from electronic case report forms of the phase III, multicenter FIL-MCL0208 trial (NCT02354313) for younger MCL patients [Cortelazzo, EHA 15]. The study enrolled 300 subjects, with median followup of 51 months. In this work we employed baseline clinical data and May '18 as survival outcomes cut-off.
For the present analysis, we started from 32 baseline features: 7 were not eligible due to number of missing values (MVs ≥40). Features with <15 MVs were imputed by median of observations. Secondly, 18 not binary variables were dichotomized, to be compared to the 7 binary ones: normal vs abnormal range or lower vs higher than a recognized cut-off value. Patients were thus split in 2 subsets, training (n=185) and validation (n=115): for the training set, only patients with no MVs were chosen.
Clustering analysis was performed to discriminate different groups of patients. Thus, we applied a recursive feature reduction, according to regression modeling, to extrapolate a restricted signature predictive of both progression free survival (PFS) and overall survival (OS). Survival analyses were done according to e-MIPI classes via both multivariate Cox and Kaplan-Maier modeling. Therefore, the e-MIPI classification was compared to known prognostic models [Hoster, Blood 08]. Finally, the signature was tested on the validation set: if any variable of the e-MIPI was missing (MVs=36, 29 and 15 for albumin - alb, Ki67 and flowcytometric peripheral blood invasion - flowpb) data mining (K-nn) technique was employed for imputation. Clustering and statistical analyses were implemented via MATLAB© and SPSS©.
Results
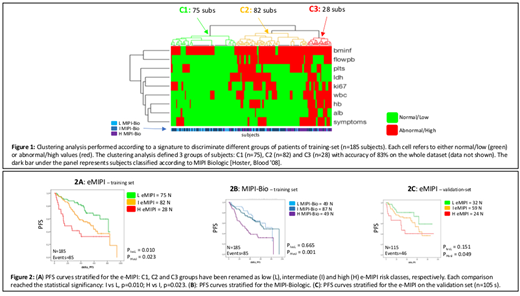
Training set: the clustering analysis allowed to define 3 groups of subjects: C1 (n=71), C2 (n=77) and C3 (n=37), showing significantly different PFS and OS. Thus, the e-MIPI index was modeled based on a signature of 9 significant features (fig 1): histologic bone marrow infiltration (bminf), flowpb, Ki67, B symptoms, platelets (plts), ldh, white blood cells (wbc), hemoglobin (hb) and alb levels. The re-clustering of the training set according to the e-MIPI confirmed the original patients clustering with 83% of accuracy. Figure 2A depicts the PFS curves stratified for the e-MIPI: C1, C2 and C3 groups have been renamed as low (L), intermediate (I) and high (H) e-MIPI risk classes, respectively. Each comparison reached the statistical significancy: I vs L, p=0.010; H vs I, p=0.023, outperforming in our series both the MIPI-St (H vs I risk, p=0.801) and MIPI-Bio (I vs L risk, p=0.665, fig. 2B) classifications.
Validation set: the e-MIPI allowed to discriminate 3 groups of subjects C1 (n=32), C2 (n=59) and C3 (n=24). Actually, the e-MIPI on the validation set (fig. 2C) confirmed the results of the training set, overall improving the MIPI-St stratification (H vs I, p=0.059 ⇒ p=0.049), even if without reaching the statistical significancy on the I vs L comparison (p=0.24 ⇒ p=0.15), due to the limited number of events in this series.
Discussion
e-Mipi is a new first prognostic index derived from hierarchical clustering. Our results indicate that this approach might allow to model engineered prognostic indexes based on comprehensive analysis of large datasets. Even if promising, it needs validation through its application to independent series of MCL patients. Additional efforts aiming at integrating biological variables in the model are ongoing.
Gaidano:Amgen: Consultancy, Honoraria; Morphosys: Honoraria; Janssen: Consultancy, Honoraria; Gilead: Consultancy, Honoraria; Roche: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria. Ladetto:Celgene: Honoraria; Sandoz: Honoraria; Jannsen: Honoraria; Roche: Honoraria; Abbvie: Honoraria; Acerta: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.